Better scientific articles — and beyond
Why I joined Curvenote
When my redesign of the scientific poster went viral (#betterposter), people asked me how we could apply the same design principles to improve scientific articles. The answer is easy to explain, but hard to implement: make articles more machine-readable than HTML first, and then you can wrap whatever interfaces you want on them — create a universe of designs for different purposes 🎨.
Now, joining Curvenote as full-time UX lead 💼, I’m super excited that I get to help actually implement this vision, and help improve the user experience of scientific articles, peer review, and scientific publishing.

Part 1: The dream¶
A scientific article should adapt to your current information need¶
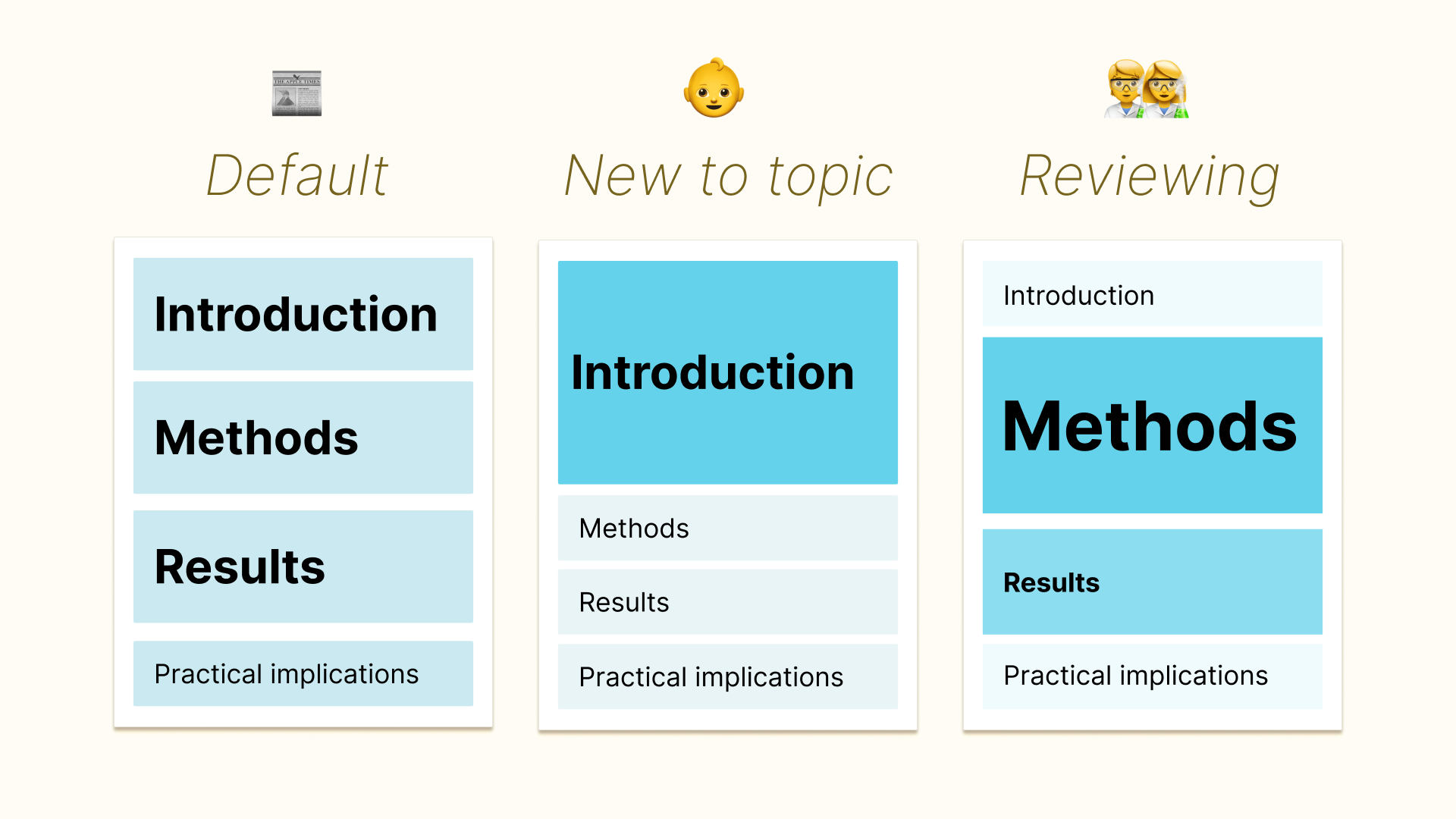
Figure 2:And this assumes you’re stuck in a single paper.
New to the topic? The article should give you a deeper introduction. Working in a company? You should be able to find the ‘practical implications’ section faster. Doing a replication study? The article’s methods should be presented as a step-by-step.
💎 It’s not about articles — it’s about that piece of insight within an article that you actually want¶
Scientists don’t always skim read articles start-to-finish 👁️. They are often interested in a particular tiny piece of the study 💎. How did they measure X? What does the effect of X on Y look like over time?
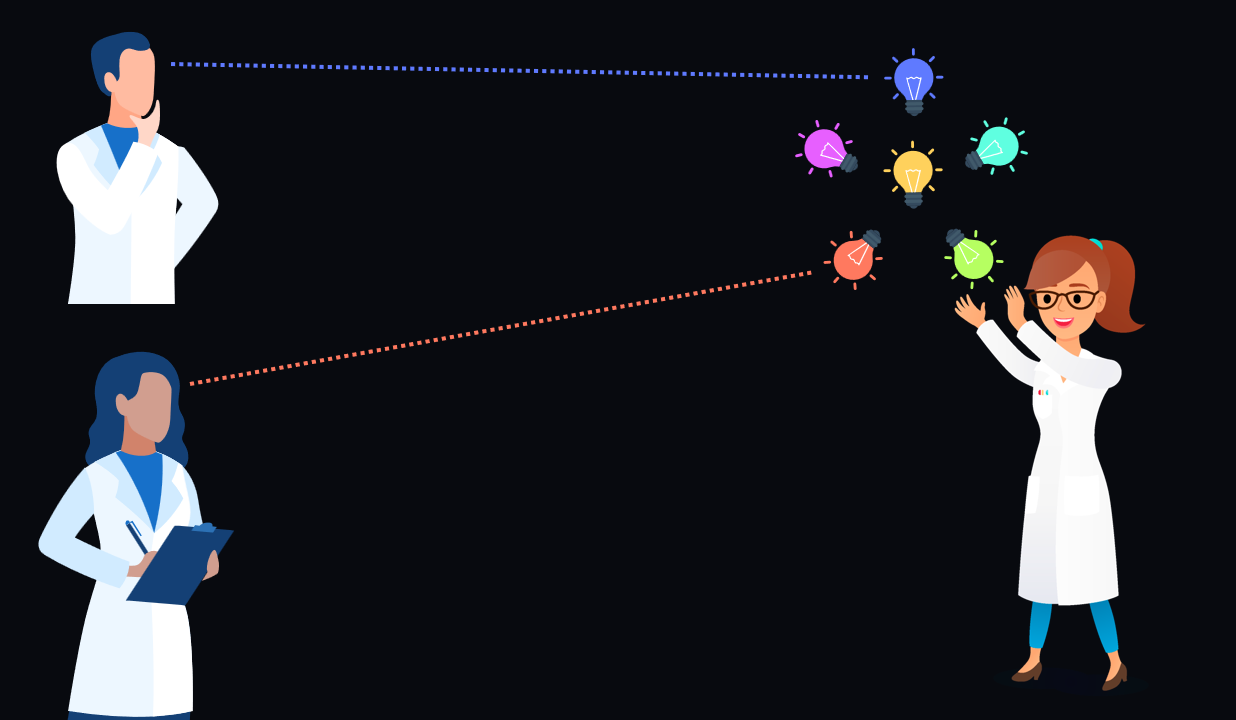
Figure 3:A single article is a bundle of individual insights. Readers may just want one particular insight in the bundle.
The goal of scientific publishing is to get the scientist to the particular piece of information 💎 they’re looking for in less than 400ms ⏱️ (called the Doherty Threshold). Scrolling through a bunch of 40 page PDF files 📄 to find the two sentences that answer your question is usually slower than 400ms.
🧱 You want to extract chunks of information across articles¶
Science is published in a single article, but scientific insight is discovered by looking across articles. If you’re trying to understand how to measure a concept, you’re real question isn’t “How did they measure X in this study?” Your real question is “How did they measure X in all studies?” To extract and display that information easily, applications need those individual chunks 🧱 of info marked up in a way that’s easy for machines to read.
♾️ A million creative ways to explore science¶
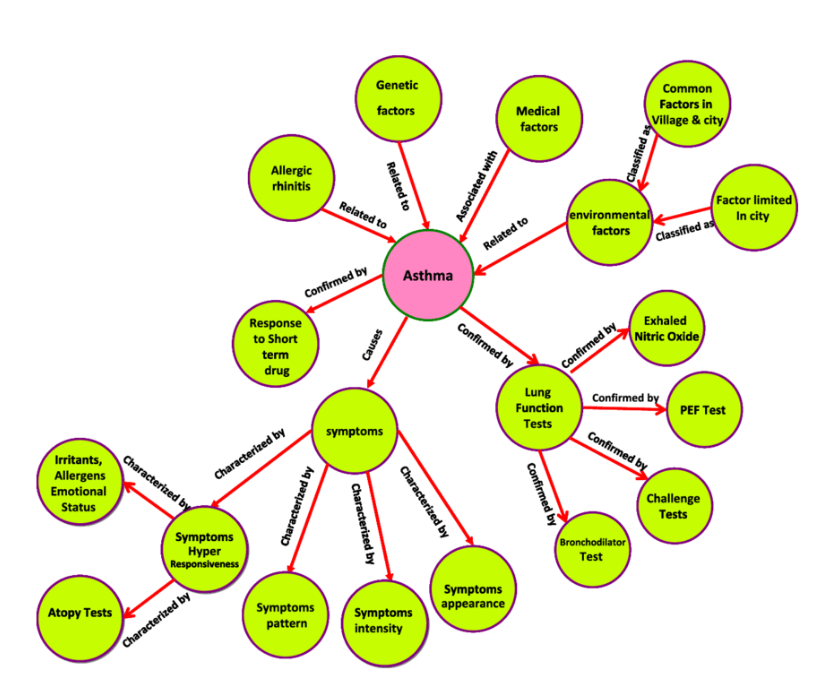
Figure 4:A network visualization of Asthma’s symptoms and risk factors
If you can get scientific articles published in a format where every important piece of the article is a machine-readable chunk, you can do anything design-wise. Every cool way you’ve seen science displayed becomes possible to apply to every article. A few examples:
- 🪄 Switch an article’s entire layout in seconds
- 🕳️🐇 Click rabbit-hole links where you can dive into cited studies without leaving the page
- Generate population-specific effect summaries
- And of course everything HTML can do: Videos, executable code, interactive charts, Google-ability.
…any summary layout that would help you do your science, you can build.
If we can get scientific articles into a chunked, machine-readable format.
Part 2: Making this happen is hard, for two reasons¶
Figure 5:Word and PDF files are less machine readable than HTML; we need a format for science that is MORE machine-readable.
Right now, with next-generation science tools like Elicit.org, Iris.ai, and Scholarcy, you’re seeing the best that 🤖 AI algorithms like GPT can do to extract scientific information from PDF files that aren’t designed for machines. These tools would become at least ten-zillion times more helpful if the format of scientific articles wasn’t actively fighting their efforts.
🤖 We need a common markup language for scientific articles¶
Figure 6:Clearly marking up the abstract paragraph in a machine-readable way makes Google Scholar Bot puke rainbows.
To get scientific articles converted into machine-readable chunks, we need a common markup language (more specific to science than HTML) for the robots to read. JATS4R is close to this, right now converting your Word doc to JATS4R costs them thousands of 💵, which they have to pass along in subscription fees and APC charges.
We want you — the paper author 👨🔬 — to be able to write your articles in a machine-readable format first, because then everything after your draft (review, publishing) gets way, way more efficient and cheaper.
📝 We need a WYSIWYG text editor so you can write papers in machine-readable markup¶
Figure 7:A typical WYSIWYG text editor that actually produces HTML.
When you use Webflow or Wordpress, you’re composing a document in HTML without code. Having the perfect markup language for scientific articles isn’t enough if nobody can use it besides publishers. We need a scientific authoring tool that feels like that Word but writes the scientific markup. And that lets you, the author, mark up things like your author affiliation so you don’t have to type them over and over again.
Part 3: Good news — it’s done¶
🤖 Meet MyST, the open-source markup language for scientific articles¶

Figure 8:MyST - An open-source markup language for scientific articles that’s enabling a new wave of scientific publishing innovations. See the MyST Documentation.
The open-source MyST Markdown language (see #WhatIsMyST on Twitter), which Curvenote donates a lot of development time to, is easier to author in, designed for scientific articles, and (crucially) converts instantly between all the other scientific document formats (JATS4R, HTML, Word). And it has incredible features for scientists, like you can copy-paste a DOI URL and auto-import a reference.
📝 Meet Curvenote, the WYSIWYG text editor you can use to write machine-readable scientific articles¶
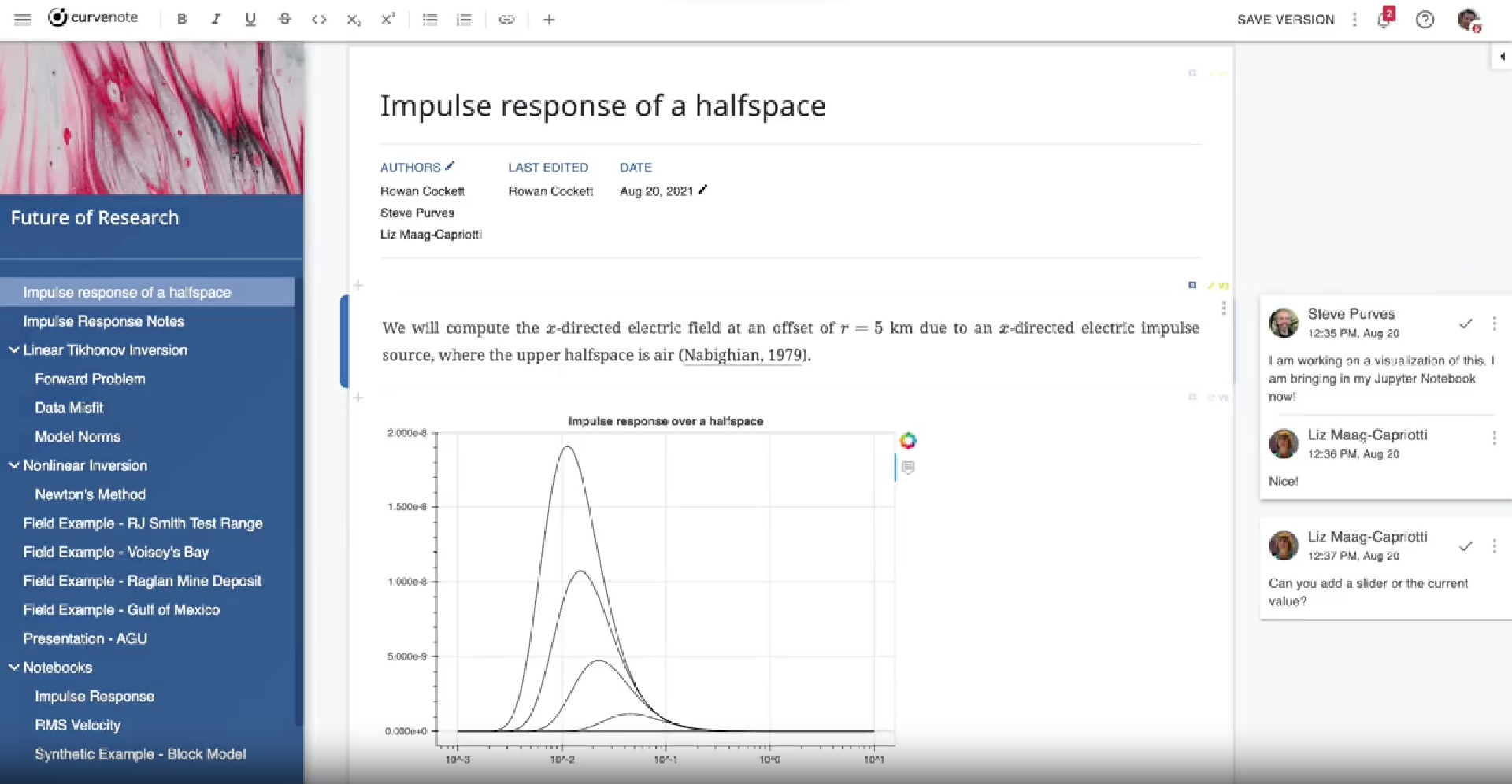
Figure 9:Curvenote: Feels like word, exports machine-readable MyST Markdown for better scientific articles.
The Curvenote WYSIWYG editor lets you write in an MS-Word like interface with features just for scientists, that can be exported as open-source MyST Markdown, JATS, , PDF, Word — whatever you want. It even lets you compose scientific articles in reusable chunks.
With these two tools alone as the core, we can start creating all-new interfaces for scientific publishing.
👨💻 Curvenote has the team to pull this off¶
When I first met the Curvenote team, my first impression was that each of them was a visionary in their own right, and exactly team I always pictured science needing to solve this problem. I’m so excited I get to join this team and take a shot at solving the hardest design problems in science.
🔮 Better articles are only the beginning¶
Since Curvenote has already built the core (a markup language and editor), we’re now focused on the million amazing innovations you can make when articles start life as machine-readable (vs PDFs). We’re already working on concepts for formless article submission systems (no more filling-out 24 author names), crazy-streamlined peer review, and links between scientific articles that are more useful even than standard web links.
But we’ll need all of your help to get the designs right, so please HMU on Bluesky and let me know about all your hopes, dreams, and frustrations for scientific publishing and distribution. My job is to make your job easier. 😊